Thrombin ligands for pharmacophore extraction
from collections import defaultdict
import os
import pandas as pd
from rdkit import Chem
from IPython.display import SVG
import openpharmacophore as oph
We want to get ligand based pharmacophores for thrombin.
We’ll start by loading a set pf thrombin ligands and generating conformers for each. Then we will store the conformers on a file to avoid needing to generate new conformers each time the notebooks is ran.
lig_df = pd.read_csv("../data/thrombin.csv")
lig_df
| Smiles | PDBID | LigID | |
|---|---|---|---|
| 0 | [H]/N=C(\C1CCC(CC1)CNC(=O)[C@@H]2C=C(CN3N2C(=O... | 1C4V | IH2 |
| 1 | CN[C@H](Cc1ccccc1)C(=O)N2CCC[C@H]2C(=O)NCC3CCC... | 1TOM | MIN |
| 2 | c1ccc(cc1)S(=O)(=O)CCN2C(=O)N3CC=C[C@H](N3C2=O... | 1D6W | 00R |
| 3 | c1ccc(cc1)S(=O)(=O)CCN2C(=O)N3CC=C[C@H](N3C2=O... | 1D91 | 00P |
| 4 | [H]/N=C(/c1ccc(cc1)C[C@H](C(=O)N2CCCCC2)NC(=O)... | 1DWD | MID |
| 5 | [H]/N=C(\c1ccc2c(c1)cc([nH]2)C(=O)N3CCC(CC3)Cc... | 1D4P | BPP |
| 6 | CCC1CCN(CC1)C(=O)[C@H](CCCNC(=[NH2+])N)NS(=O)(... | 1FPC | 0ZI |
def load_ligands():
smiles = lig_df["Smiles"].tolist()
return oph.load_ligands(smiles, form="smi")
conf_file = "../data/thrombin_ligands.sdf"
if not os.path.isfile(conf_file):
ligands = load_ligands()
else:
ligands = oph.load(conf_file)
print("Number of conformers ", [l.n_conformers for l in ligands])
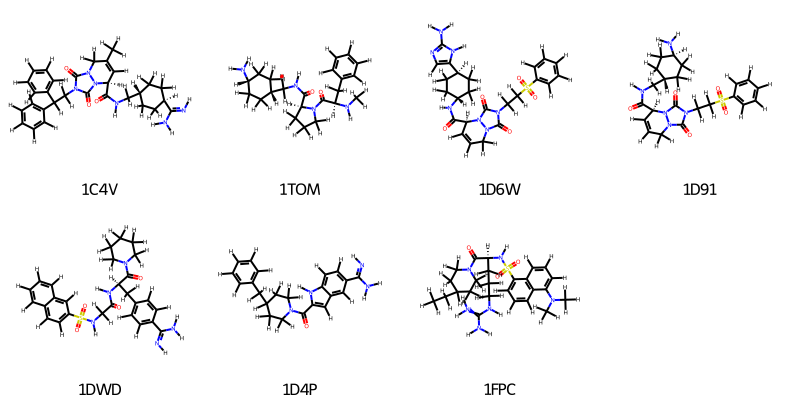
oph.draw_ligands(ligands, n_per_row=4, legends=[l.to_rdkit().GetProp("_Name") for l in ligands])
Number of conformers [176, 174, 166, 147, 176, 40, 286]
Now we generate conformers for each ligand. The argument n_confs is set to -1 so that the conformer generator creates and “ideal” number of conformers for each ligand.
for lig in ligands:
if not lig.has_conformers:
lig.add_hydrogens()
lig.generate_conformers(n_confs=-1)
for ii, lig in enumerate(ligands):
print(f"Ligand {ii} has {lig.n_conformers} conformers")
Ligand 0 has 176 conformers
Ligand 1 has 174 conformers
Ligand 2 has 166 conformers
Ligand 3 has 147 conformers
Ligand 4 has 176 conformers
Ligand 5 has 40 conformers
Ligand 6 has 286 conformers
# Save conformers to file
if not os.path.isfile(conf_file):
writer = Chem.SDWriter(conf_file)
for lig in ligands:
lig_rdkit = lig.to_rdkit()
for conf in range(lig.n_conformers):
writer.write(lig_rdkit, confId=conf)
writer.close()
Analyizing chem feats
def chem_feats_df():
n_feats = defaultdict(list)
names = []
for lig in ligands:
names.append(lig.to_rdkit().GetProp("_Name"))
for feat_type, ind_list in lig.feat_ind.items():
n_feats[feat_type].append(len(ind_list))
df = pd.DataFrame.from_dict(n_feats)
df["ligand"] = names
df.set_index("ligand", drop=True, inplace=True)
df["total"] = df.sum(axis=1)
return df
for lig in ligands:
lig.get_chem_feats(0)
chem_feats_df()
| aromatic ring | hydrophobicity | negative charge | positive charge | hb acceptor | hb donor | total | |
|---|---|---|---|---|---|---|---|
| ligand | |||||||
| 1C4V | 3 | 2 | 0 | 1 | 3 | 3 | 12 |
| 1TOM | 1 | 3 | 0 | 2 | 4 | 3 | 13 |
| 1D6W | 3 | 2 | 0 | 0 | 6 | 3 | 14 |
| 1D91 | 2 | 2 | 0 | 1 | 6 | 2 | 13 |
| 1DWD | 3 | 2 | 0 | 1 | 5 | 4 | 15 |
| 1D4P | 3 | 0 | 0 | 1 | 2 | 3 | 9 |
| 1FPC | 2 | 4 | 0 | 2 | 3 | 4 | 15 |
drawing = oph.draw_ligands_chem_feats(ligands, lig_size=(300, 280))
SVG(drawing.GetDrawingText())
Finding common pharmacophores
pharma_file = "../data/thrombin_pharmacophores.mol2"
import time
start = time.time()
n_points = [3, 4, 5, 6]
min_actives = [4, 5, 6]
if not os.path.isfile(pharma_file):
pharmacophores = []
for ii in range(len(n_points)):
for jj in range(len(min_actives)):
pnts = n_points[ii]
actives = min_actives[jj]
pharma = oph.LigandBasedPharmacophore(ligands)
pharma.extract(
n_points=pnts, min_actives=actives, max_pharmacophores=10
)
print(f"Num points: {pnts}. Min actives: {actives}. Found {len(pharma)} pharmacophores")
if len(pharma) > 0:
for p in pharma.pharmacophores:
p.props["min_actives"] = actives
pharmacophores.extend(pharma.pharmacophores)
else:
pharmacophores = oph.pharmacophore_reader.read_mol2(pharma_file)
print(f"Found {len(pharmacophores)} pharmacopohores")
Found 63 pharmacopohores
end = time.time() - start
print(f"{end} seconds")
0.22133302688598633 seconds
# Save pharmacophores to file
if not os.path.isfile(pharma_file):
oph.pharmacophore_writer.save_mol2(pharmacophores, pharma_file)
def pharmacophore_df():
rows = {}
ii = 0
for pharma in pharmacophores:
feat_str = ""
# Only add the top scoring pharmacophore
for point in pharma:
feat_str += point.short_name
n_pnts = len(pharma)
n_actives = int(pharma.props["min_actives"])
rows[ii] = [n_pnts, n_actives, feat_str, pharma.ref_mol, pharma.score]
ii += 1
df = pd.DataFrame.from_dict(
rows,
orient="index",
columns=["Number of points", "Number of actives", "Features", "Reference ligand", "Score"]
)
return df
df = pharmacophore_df()
df
| Number of points | Number of actives | Features | Reference ligand | Score | |
|---|---|---|---|---|---|
| 0 | 3 | 4 | HPR | 0 | 0.947654 |
| 1 | 3 | 4 | ADR | 2 | 0.957363 |
| 2 | 3 | 4 | AHP | 1 | 0.959299 |
| 3 | 3 | 4 | ADH | 3 | 0.948446 |
| 4 | 3 | 4 | AHR | 1 | 0.969500 |
| ... | ... | ... | ... | ... | ... |
| 58 | 5 | 4 | AADDP | 0 | 0.301428 |
| 59 | 5 | 4 | AADHP | 6 | 0.248704 |
| 60 | 5 | 4 | AAADP | 4 | 0.202498 |
| 61 | 5 | 5 | AADHR | 6 | 0.241077 |
| 62 | 5 | 5 | AADHP | 4 | 0.139471 |
63 rows × 5 columns
indices = []
for pnts in n_points:
for actives in min_actives:
filter_ = (df["Number of points"] == pnts) & (df["Number of actives"] == actives)
scores = df[filter_]["Score"]
if not scores.empty:
indices.append(scores.idxmax())
df = df.iloc[indices]
df.set_index("Number of points", drop=True, inplace=True)
df
| Number of actives | Features | Reference ligand | Score | |
|---|---|---|---|---|
| Number of points | ||||
| 3 | 4 | AHR | 1 | 0.969500 |
| 3 | 5 | AHR | 1 | 0.969500 |
| 3 | 6 | AHR | 1 | 0.969500 |
| 4 | 4 | ADHP | 3 | 0.830411 |
| 4 | 5 | ADPR | 3 | 0.807592 |
| 4 | 6 | ADPR | 3 | 0.671040 |
| 5 | 4 | ADHPR | 3 | 0.766332 |
| 5 | 5 | AADHR | 6 | 0.241077 |
# Get the highest scoring pharmacophores and the one with most features
top_score = max((p for p in pharmacophores), key=lambda x: x.score)
most_feats = max((p for p in pharmacophores), key=lambda x: len(x))
Visualizing the pharmacophores
viewer_1 = oph.Viewer()
ligand = ligands[top_score.ref_mol]
viewer_1.add_components([top_score, ligand])
print(f"Score: {top_score.score}")
print(f"Num points: {len(top_score)}")
Score: 0.9695001830941523
Num points: 3
viewer_1.show(struct=top_score.ref_struct)
viewer_2 = oph.Viewer()
ligand = ligands[most_feats.ref_mol]
viewer_2.add_components([most_feats, ligand])
print(f"Score: {most_feats.score}")
print(f"Num points: {len(most_feats)}")
Score: 0.4623265470544659
Num points: 5
viewer_2.show(struct=most_feats.ref_struct)