Virtual Screening with a pharmacophore
import pyunitwizard as puw
import openpharmacophore as oph
from openpharmacophore import mol_db
We want to screen a database of molecules with a pharmacophore to find potential matches. We start by defining our pharmacophore model
pharmacophore = oph.Pharmacophore([
oph.PharmacophoricPoint(
feat_type="hb acceptor",
center=puw.quantity([2.35, 1.94, -0.68], "angstroms"),
radius=puw.quantity(1.0, "angstroms"),
),
oph.PharmacophoricPoint(
feat_type="hydrophobicity",
center=puw.quantity([1.57, -2.53, 2.15], "angstroms"),
radius=puw.quantity(1.0, "angstroms"),
),
oph.PharmacophoricPoint(
feat_type="aromatic ring",
center=puw.quantity([6.19, 1.34, -1.36], "angstroms"),
radius=puw.quantity(1.0, "angstroms"),
),
])
We will create a database of molecules loading them from different files
file1 = "../data/screening/ligands_1.smi"
file2 = "../data/screening/ligands_2.smi"
! cat {file1}
Smiles Name
[H]/N=C(\C1CCC(CC1)CNC(=O)[C@@H]2C=C(CN3N2C(=O)N(C3=O)CC(c4ccccc4)c5ccccc5)C)/N IH2
CN[C@H](Cc1ccccc1)C(=O)N2CCC[C@H]2C(=O)NCC3CCC(CC3)N MIN
c1ccc(cc1)S(=O)(=O)CCN2C(=O)N3CC=C[C@H](N3C2=O)C(=O)NC4CCC(CC4)c5cnc([nH]5)N 00R
c1ccc(cc1)S(=O)(=O)CCN2C(=O)N3CC=C[C@H](N3C2=O)C(=O)NCC4CCC(CC4)N 00P
[H]/N=C(/c1ccc(cc1)C[C@H](C(=O)N2CCCCC2)NC(=O)CNS(=O)(=O)c3ccc4ccccc4c3)\N MID
! cat {file2}
Smiles Name
[H]/N=C(\c1ccc2c(c1)cc([nH]2)C(=O)N3CCC(CC3)Cc4ccccc4)/N BPP
CCC1CCN(CC1)C(=O)[C@H](CCCNC(=[NH2+])N)NS(=O)(=O)c2cccc3c2cccc3N(C)C 0ZI
C[C@H]1CN2c3c(cc(cc3NC2=S)Cl)CN1CC=C(C)C TB9
CC(C)c1c(n(c(n1)COC(=O)N)Cc2ccncc2)Sc3cc(cc(c3)Cl)Cl S11
CC(C)OC(=O)N1c2cc(ccc2NC(=S)[C@@H]1CSC)OC HBY
db = mol_db.MolDB()
db.from_file_list([file1, file2], header=True)
We can now create a virtual screening object to which will pass our pharmacophore and our molecule db
screener = oph.VirtualScreening([pharmacophore])
screener.screen(db)
print(f"{len(screener.matches[0])} molecule matched the pharmacophore")
print(f"{screener.fails[0]} molecule failed to match the pharmacophore")
4 molecule matched the pharmacophore
6 molecule failed to match the pharmacophore
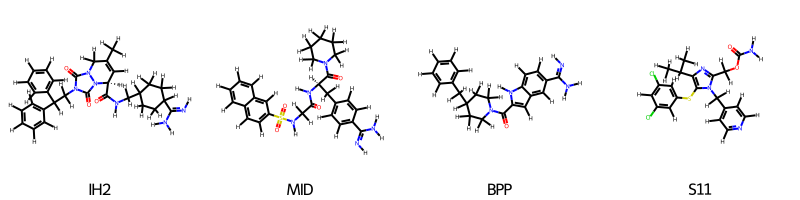
We inspect the molecules that matched the pharmachore along with their alignment scores (RMSD)
ligands = [m.ligand for m in screener.matches[0]]
legends = [
m.ligand.to_rdkit().GetProp("_Name") for m in screener.matches[0]
]
scores = [m.rmsd for m in screener.matches[0]]
print("RMSDs", scores)
oph.draw_ligands(
ligands=ligands,
n_per_row=4,
legends=legends
)
RMSDs [0.8177257515854292, 0.8517769586370709, 0.7687604258063847, 0.9831765173894478]
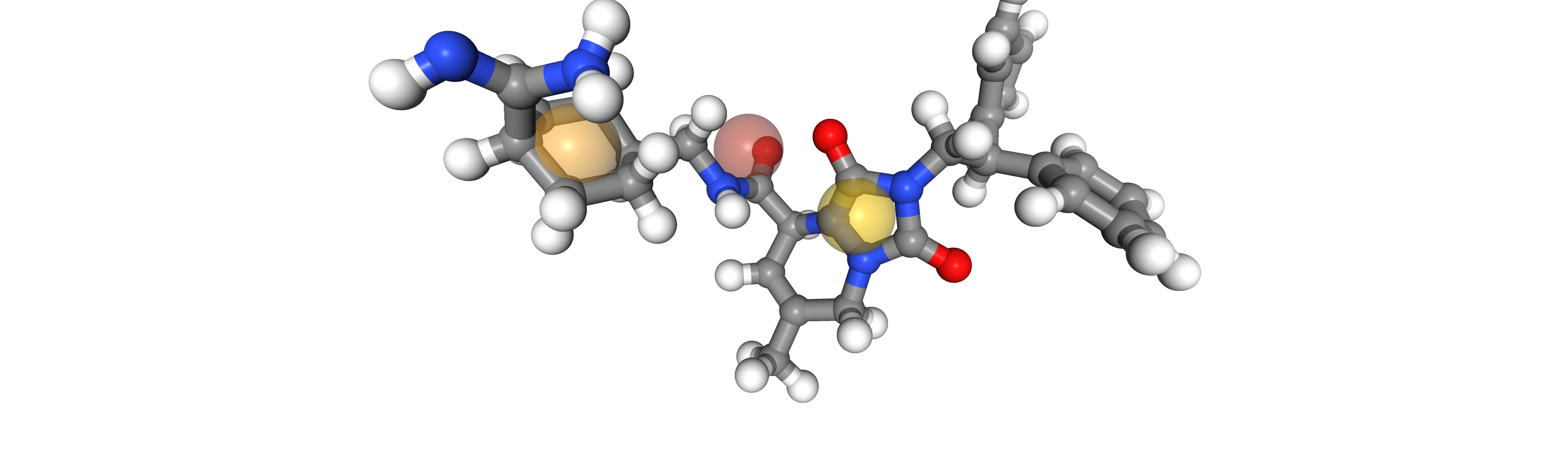
We visualize the aligned ligands and the pharmacophore
idx = 0
viewer = oph.Viewer()
viewer.add_components([ligands[idx], pharmacophore])
viewer.show()