Binding site of a protein-ligand complex
In this notebook we explore the binding site of two different protein ligand-complexes.
We’ll start by loading the complexes structures from a pdb file. We then proceed to prepare the ligand and the protein so we can extract and visualize the binding site of the protein
import openpharmacophore as oph
PDB 3BBH
We start with the structure with PDB ID 3BBH. This file was already pretreated, so the protein contains hydrogens, and there is only a single chain.
protein = oph.load("../../data/3bbh_hyd.pdb")
print(f"Has hydrogens: {protein.has_hydrogens}")
print(f"Has ligand: {protein.has_ligands}")
Has hydrogens: <bound method Protein.has_hydrogens of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f068c023910>>
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f068c023910>>
A protein may contain one or multiple ligands. To extract one we need to know its id. We can view
the id of all the ligans in the protein by calling the attribute ligand_ids
lig_ids = protein.ligand_ids()
print(lig_ids)
['SFG:B']
ligand = protein.get_ligand(lig_ids[0])
ligand.draw()
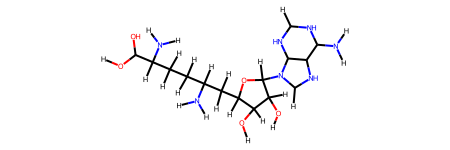
We fix the ligand. Its bond orders are correct now
ligand.fix_bond_order(
smiles="c1nc(c2c(n1)n(cn2)[C@H]3[C@@H]([C@@H]([C@H](O3)C[C@H](CC[C@@H](C(=O)O)N)N)O)O)N"
)
ligand.draw()
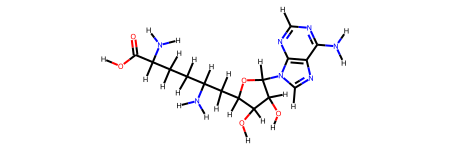
Now we can remove the ligand from the protein
protein.remove_ligand(lig_ids[0])
print(f"Has ligand: {protein.has_ligands}")
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f068c023910>>
We create a binding site to obtain the receptor chemical features
bsite = oph.ComplexBindingSite(protein, ligand)
# Extract chemical features and visualize them
receptor_feats = bsite.get_chem_feats(frame=0)
ligand_feats = ligand.get_chem_feats(conf_ind=0)
print(f"Found {len(receptor_feats)} chemical features in receptor")
print(f"Found {len(ligand_feats)} chemical features in ligand")
Found 492 chemical features in receptor
Found 20 chemical features in ligand
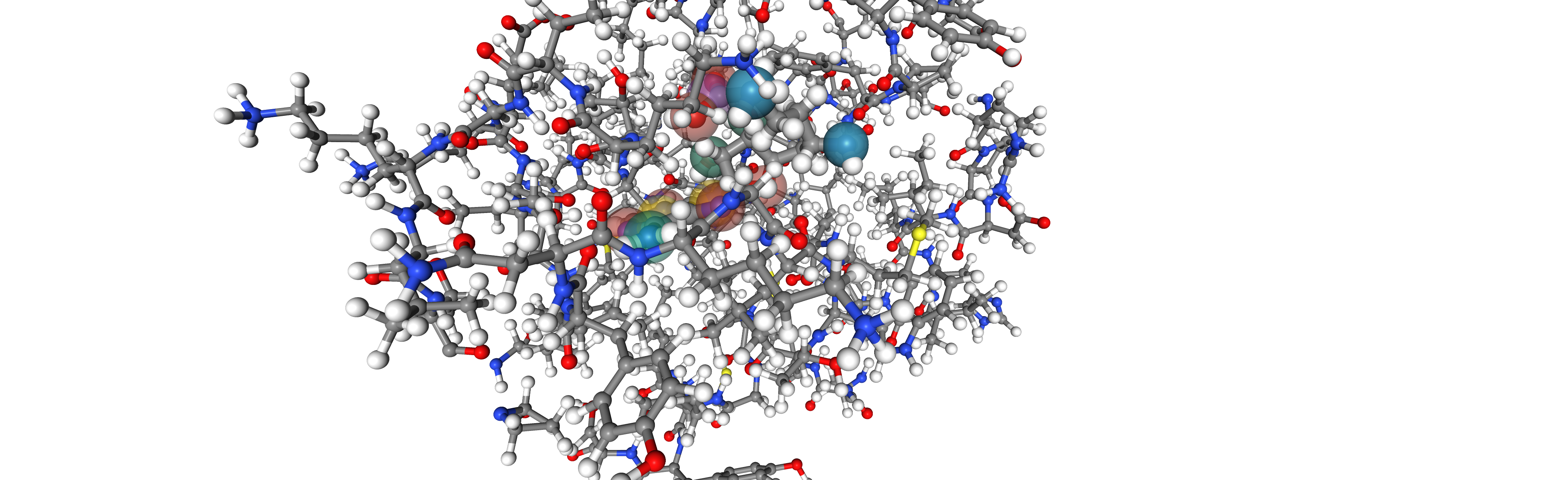
Finally we visualize the binding site with the chemical
viewer = oph.Viewer()
viewer.add_components([bsite, ligand, ligand_feats])
view = viewer.show()
viewer.set_protein_style("ball+stick")
view
PDB 1M7W
protein_2 = oph.load("../../data/1m7w_A_chain.pdb")
print(f"Has hydrogens: {protein_2.has_hydrogens}")
print(f"Has ligand: {protein_2.has_ligands}")
Has hydrogens: <bound method Protein.has_hydrogens of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f0650baf210>>
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f0650baf210>>
lig_ids = protein_2.ligand_ids()
print(lig_ids)
['DAO:B']
ligand_2 = protein_2.get_ligand(lig_ids[0])
ligand_2.draw()
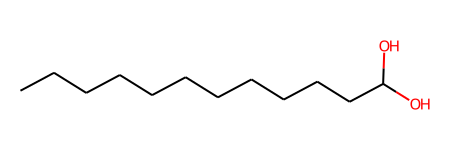
ligand_2.fix_bond_order(smiles="CCCCCCCCCCCC(=O)O")
ligand_2.add_hydrogens()
ligand_2.draw()
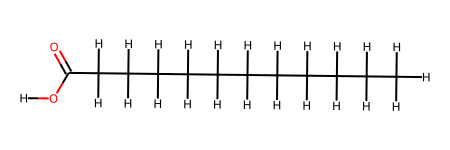
protein_2.remove_ligand(lig_ids[0])
print(f"Has ligand: {protein_2.has_ligands}")
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f0650baf210>>
protein_2.add_hydrogens()
print(f"Has hydrogens: {protein_2.has_hydrogens}")
Has hydrogens: <bound method Protein.has_hydrogens of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f0650baf210>>
bsite_2 = oph.ComplexBindingSite(protein_2, ligand_2)
receptor_feats_2 = bsite_2.get_chem_feats(frame=0)
ligand_feats_2 = ligand_2.get_chem_feats(conf_ind=0)
print(f"Found {len(receptor_feats_2)} chemical features in receptor")
print(f"Found {len(ligand_feats_2)} chemical features in ligand")
Found 652 chemical features in receptor
Found 9 chemical features in ligand
viewer_2 = oph.Viewer()
viewer_2.add_components([bsite_2,ligand_2, ligand_feats_2])
view_2 = viewer_2.show()
viewer_2.set_protein_style("ball+stick")
view_2
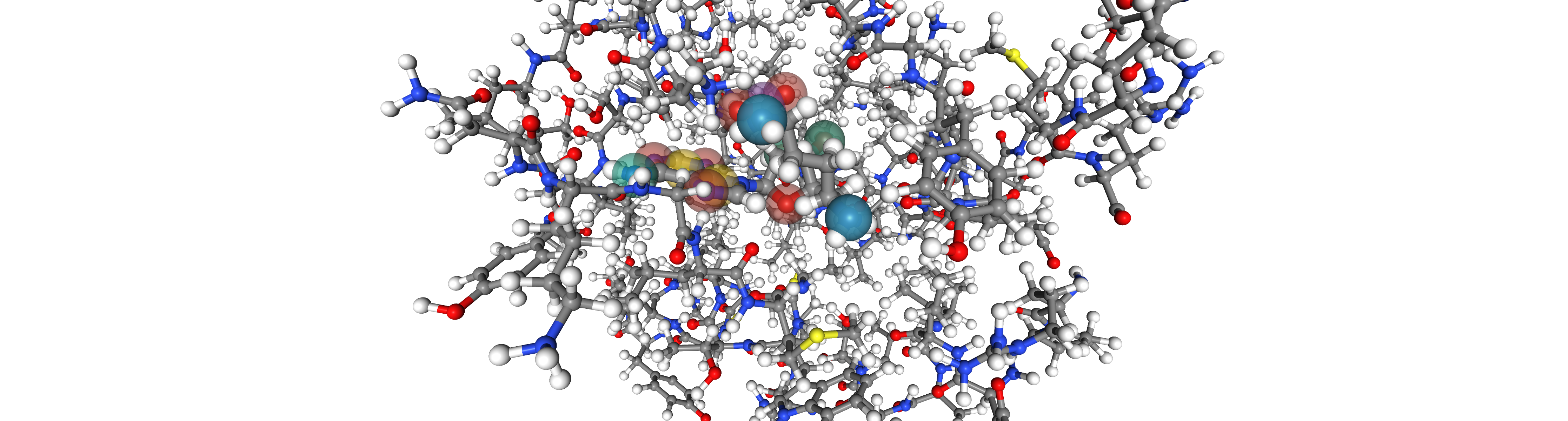
view and view_2 display an interactive widget. For simplicity an image is presented in the documentation.
