Hydrophobic interactions
import openpharmacophore as oph
pdb_path = "../../data/1m7w_A_chain.pdb"
protein = oph.load(pdb_path)
print(f"Has hydrogens: {protein.has_hydrogens()}")
print(f"Has ligand: {protein.has_ligands()}")
Has hydrogens: False
Has ligand: True
lig_ids = protein.ligand_ids()
print(lig_ids)
['DAO:B']
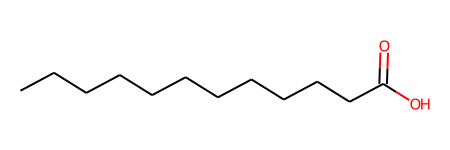
smiles = oph.smiles_from_pdb_id(lig_ids[0])
smiles
'CCCCCCCCCCCC(=O)O'
ligand = protein.get_ligand(lig_ids[0])
ligand.fix_bond_order(smiles=smiles)
ligand.draw()
protein.remove_ligand(lig_ids[0])
print(f"Has ligand: {protein.has_ligands}")
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7f2454e8c990>>
bsite = oph.ComplexBindingSite(protein, ligand)
pharmacophore = oph.LigandReceptorPharmacophore(bsite, ligand)
pharmacophore.extract(feat_types=["hydrophobicity"])
print(f"Number of pharmacophoric points {len(pharmacophore[0])}")
for p in pharmacophore[0]:
print(p)
Number of pharmacophoric points 5
PharmacophoricPoint(feat_type=hydrophobicity; center=(64.61, 33.63, 10.33); radius=1.0)
PharmacophoricPoint(feat_type=hydrophobicity; center=(64.0, 33.95, 8.65); radius=1.0)
PharmacophoricPoint(feat_type=hydrophobicity; center=(62.71, 27.31, 13.27); radius=1.0)
PharmacophoricPoint(feat_type=hydrophobicity; center=(64.26, 29.96, 13.92); radius=1.0)
PharmacophoricPoint(feat_type=hydrophobicity; center=(65.22, 31.7, 13.23); radius=1.0)
viewer = oph.Viewer()
viewer.add_components([bsite, ligand, pharmacophore[0]])
view = viewer.show()
viewer.set_protein_style("ball+stick")
view
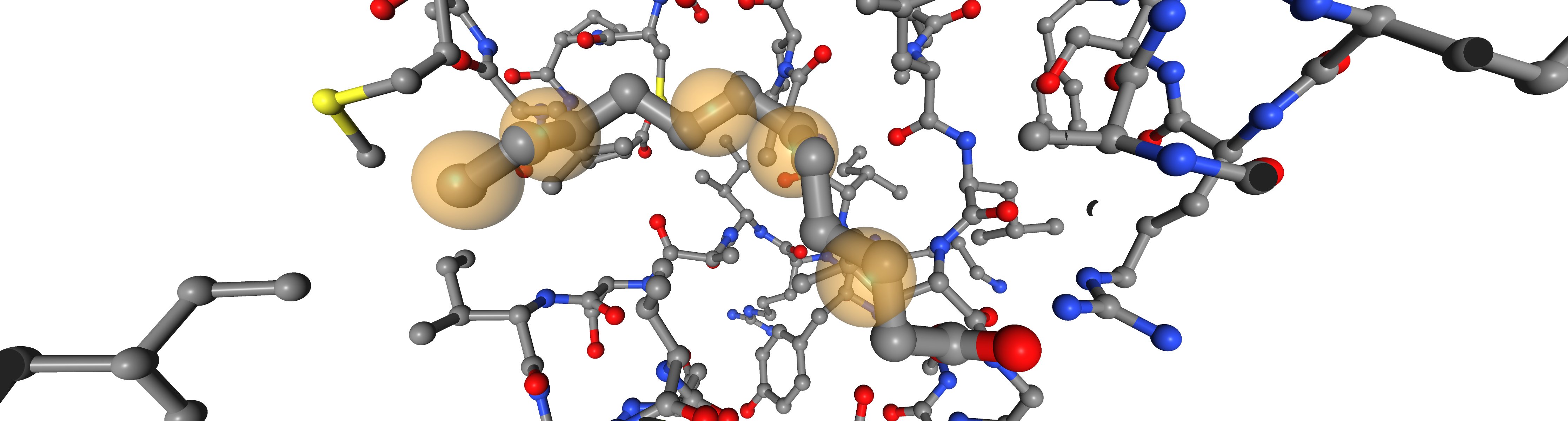
Note:
viewer.show() displays an interactive widget. For simplicity an image is presented in the documentation.
