import openpharmacophore as oph
import pyunitwizard as puw
import numpy as np
Er-alpha
Pharmacophore of the protein-ligand complex of estrogen receptor with estradiol.
protein = oph.load("../data/er_alpha_A_chain.pdb")
print(f"Protein has {protein.n_atoms} atoms")
# We know that the file contains a single ligand
lig_ids = protein.ligand_ids()
print(lig_ids)
Protein has 2010 atoms
['EST:B']
We obtain the smiles of the ligand. Necessary to fix its bond order later
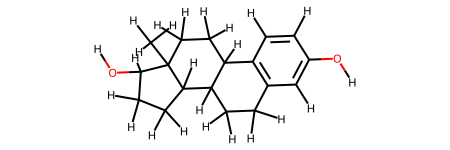
smiles = oph.smiles_from_pdb_id(lig_ids[0])
smiles
'C[C@]12CC[C@@H]3c4ccc(cc4CC[C@H]3[C@@H]1CC[C@@H]2O)O'
We extract the ligand and fix its bond order and add hydrogens
ligand = protein.get_ligand(lig_ids[0])
ligand.fix_bond_order(smiles=smiles)
ligand.add_hydrogens()
ligand.draw()
protein.remove_ligand(lig_ids[0])
print(f"Has ligand: {protein.has_ligands}")
Has ligand: <bound method Protein.has_ligands of <openpharmacophore.molecular_systems.protein.Protein object at 0x7fe2c8e08650>>
We add hydrogens to the protein
protein.add_hydrogens()
print(f"Protein has {protein.n_atoms} atoms after adding hydrogens")
Protein has 4039 atoms after adding hydrogens
We need to extract the binding site from the protein, so we can get pharmacophoric features
bsite = oph.ComplexBindingSite(protein, ligand)
Obtaining the pharmacophore
pharmacophore = oph.LigandReceptorPharmacophore(bsite, ligand)
pharmacophore.extract()
print(f"Number of pharmacophoric points {len(pharmacophore[0])}")
for p in pharmacophore[0]:
print(p)
Number of pharmacophoric points 3
PharmacophoricPoint(feat_type=aromatic ring; center=(102.81, 16.72, 24.52); radius=1.0; direction=(0.09, 0.5, -0.86))
PharmacophoricPoint(feat_type=hydrophobicity; center=(107.21, 12.57, 22.11); radius=1.0)
PharmacophoricPoint(feat_type=hydrophobicity; center=(107.38, 12.45, 24.42); radius=1.0)
viewer = oph.Viewer()
viewer.add_components([protein, ligand, pharmacophore[0]])
viewer.show()
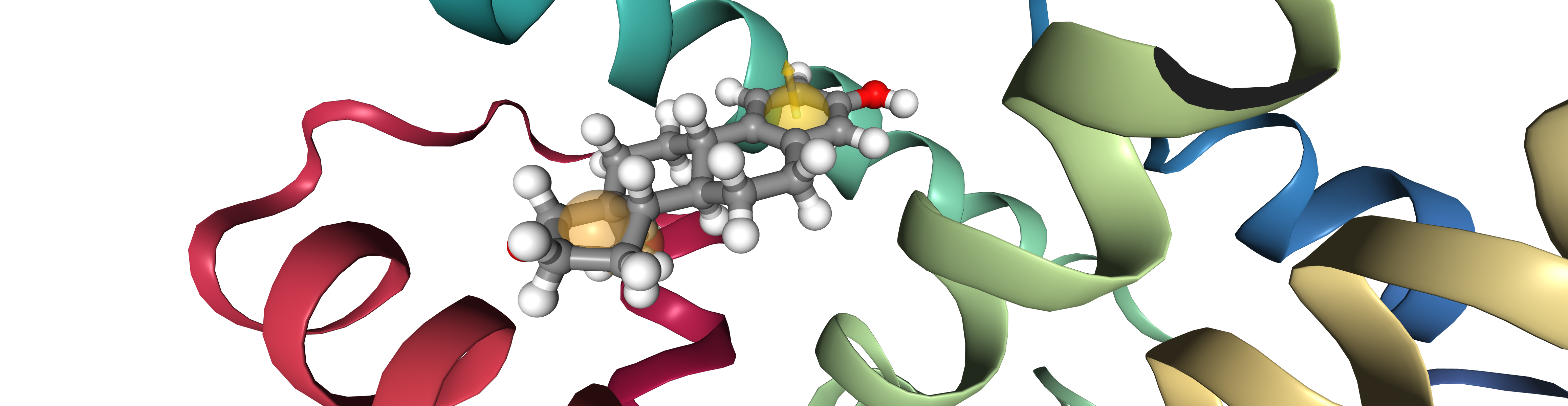
Note:
viewer.show() displays an interactive widget. For simplicity an image is presented in the documentation.
