Get non bonded potential energy#
import molsysmt as msm
msm.config.set_default_standard_units(standards=['nm', 'ps', 'K', 'mole', 'amu', 'e',
'kcal/mol', 'kcal/(mol*nm**2)', 'N', 'degrees'])
molecular_system = msm.convert(msm.systems['Barnase-Barstar']['barnase_barstar.h5msm'])
msm.info(molecular_system, element='molecule')
| index | name | type | n atoms | n groups | n components | chain index | entity index | entity name |
|---|---|---|---|---|---|---|---|---|
| 0 | BARNASE | protein | 1727 | 110 | 1 | 0 | 0 | BARNASE |
| 1 | BARSTAR | protein | 1432 | 89 | 1 | 0 | 1 | BARSTAR |
U1nb2 = msm.molecular_mechanics.get_non_bonded_potential_energy(molecular_system,
selection='molecule_name=="BARNASE"',
selection_2='molecule_name=="BARSTAR"')
U1nb2
-761.4179568126493 kilocalorie/mole
U12 = msm.molecular_mechanics.get_potential_energy(molecular_system)
U1 = msm.molecular_mechanics.get_potential_energy(molecular_system, selection='molecule_name=="BARNASE"')
U2 = msm.molecular_mechanics.get_potential_energy(molecular_system, selection='molecule_name=="BARSTAR"')
U12-U1-U2
-761.4178701978908 kilocalorie/mole
U12_groups = msm.molecular_mechanics.get_non_bonded_potential_energy(molecular_system,
selection='all in groups of group_index in [0,1,2]',
selection_2='all in groups of group_index in [100,101,102]')
U12_groups.shape
(3, 3)
U12_groups = msm.molecular_mechanics.get_non_bonded_potential_energy(molecular_system,
selection='all in groups of molecule_name=="BARNASE"',
selection_2='all in groups of molecule_name=="BARSTAR"')
U12_groups
| Magnitude | [[22.013239719215814 8.221300450611297 0.022280874019257196 ... -0.09654084065604962 0.02127382405687012 -7.069599004480168] [-0.6920593874404353 -0.21573439610847778 0.0021243699422074094 ... 0.003396681230337168 0.000630549901305581 0.15429169452441124] [0.3373296949193309 0.10278165682324031 0.0006231705499652694 ... -0.0027903635232899883 0.0009415938339087521 -0.0857830503474686] ... [17.796437097553152 7.594372984556094 0.056892006620629064 ... -0.11979981573087767 0.09337406655342922 -7.7283131688776185] [0.2754433775039973 0.09804992557253044 0.0005992375186023019 ... -0.0029858166127542233 0.0016122315627371143 -0.09203897379552428] [-0.1815851514243307 -0.2708179772926422 0.001121433469585844 ... 0.010794936812851715 -0.017838997777056533 0.5048821122878756]] |
|---|---|
| Units | kilocalorie/mole |
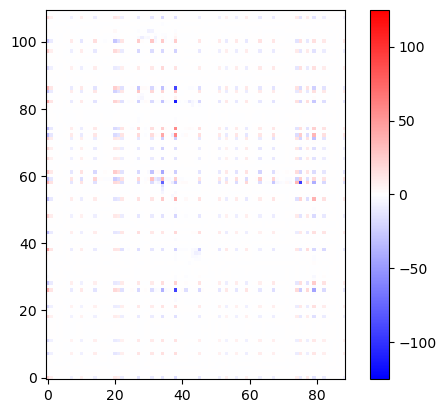
import matplotlib.pyplot as plt
plt.imshow(U12_groups, origin='lower', cmap='bwr', vmin=-125, vmax=125)
plt.colorbar()
plt.show()
/home/diego/Myopt/miniconda3/envs/MolSysMT@uibcdf_3.12/lib/python3.12/site-packages/matplotlib/cbook.py:684: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
x = np.array(x, subok=True, copy=copy)
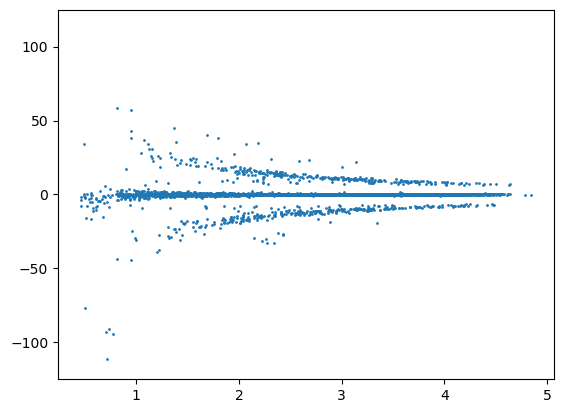
distance = msm.structure.get_distances(molecular_system, selection='all in groups of molecule_name=="BARNASE"',
selection_2='all in groups of molecule_name=="BARSTAR"')
plt.scatter(distance.flatten(), U12_groups.flatten(), s=1.0)
plt.ylim([-125.0, 125.0])
plt.show()
/home/diego/Myopt/miniconda3/envs/MolSysMT@uibcdf_3.12/lib/python3.12/site-packages/numpy/ma/core.py:2820: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
_data = np.array(data, dtype=dtype, copy=copy,
import numpy as np
U12_1_groups= U12_groups.sum(axis=1)
U12_2_groups= U12_groups.sum(axis=0)
plt.bar(np.arange(U12_1_groups.shape[0]), msm.pyunitwizard.get_value(U12_1_groups))
plt.show()
plt.bar(np.arange(U12_2_groups.shape[0]), msm.pyunitwizard.get_value(U12_2_groups))
plt.show()
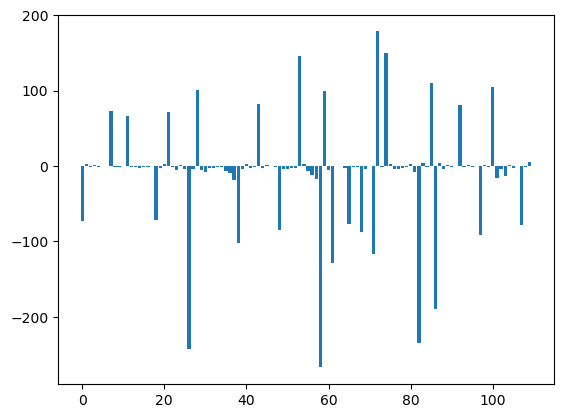
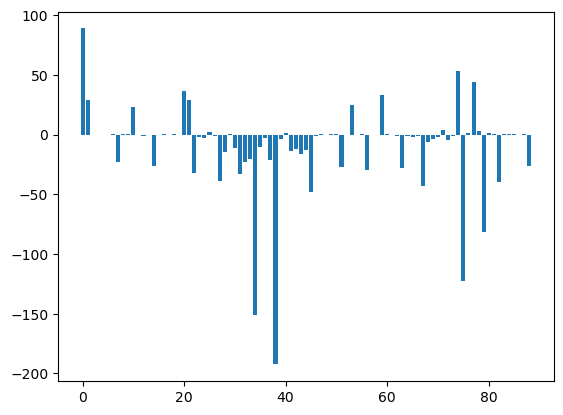
aux = [ii for ii in msm.pyunitwizard.get_value(U12_1_groups)]
aux += [ii for ii in msm.pyunitwizard.get_value(U12_2_groups)]
aux = np.array(aux)
max_abs_val = max(abs(aux.min()), abs(aux.max()))
view = msm.view(molecular_system)
view.clear()
view.add_cartoon(selection='all')
msm.thirds.nglview.set_color_by_value(view, aux, mid_value=0.0, cmap='bwr_r')
view