%load_ext autoreload
%autoreload 2
import molsysmt as msm
import numpy as np
import matplotlib.pyplot as plt
Center#
molecular_system = msm.systems['pentalanine']['traj_pentalanine.h5']
molecular_system = msm.convert(molecular_system, to_form='molsysmt.MolSys')
msm.info(molecular_system)
| form | n_atoms | n_groups | n_components | n_chains | n_molecules | n_entities | n_peptides | n_structures |
|---|---|---|---|---|---|---|---|---|
| molsysmt.MolSys | 62 | 7 | 1 | 1 | 1 | 1 | 1 | 5000 |
n_structures = msm.get(molecular_system, n_structures=True)
origin = np.zeros([n_structures, 1, 3])*msm.pyunitwizard.unit('nanometers')
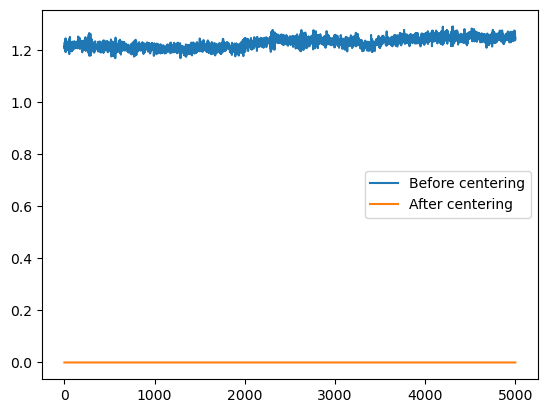
geometric_center = msm.structure.get_center(molecular_system)
plt.plot(geometric_center[:,0,0])
plt.plot(geometric_center[:,0,1])
plt.plot(geometric_center[:,0,2])
plt.show()
MOLSYSMT WARNING | UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
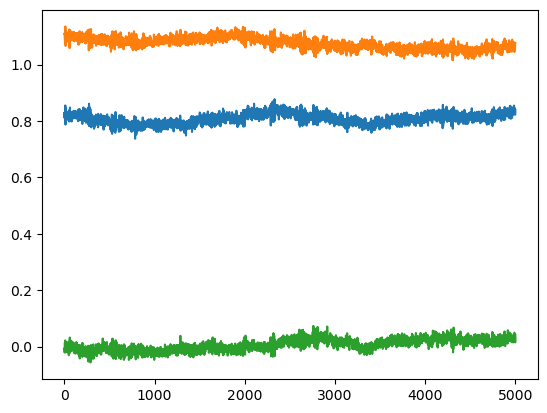
distance_before = msm.structure.get_distances(molecular_system, center_of_atoms=True, molecular_system_2=origin)
molecular_system = msm.structure.center(molecular_system)
distance_after = msm.structure.get_distances(molecular_system, center_of_atoms=True,
molecular_system_2=origin)
plt.plot(distance_before[:,0,0], label='Before centering')
plt.plot(distance_after[:,0,0], label='After centering')
plt.legend()
plt.show()
MOLSYSMT WARNING | UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.